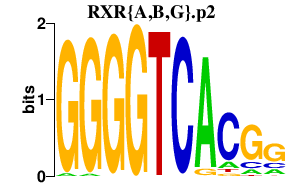
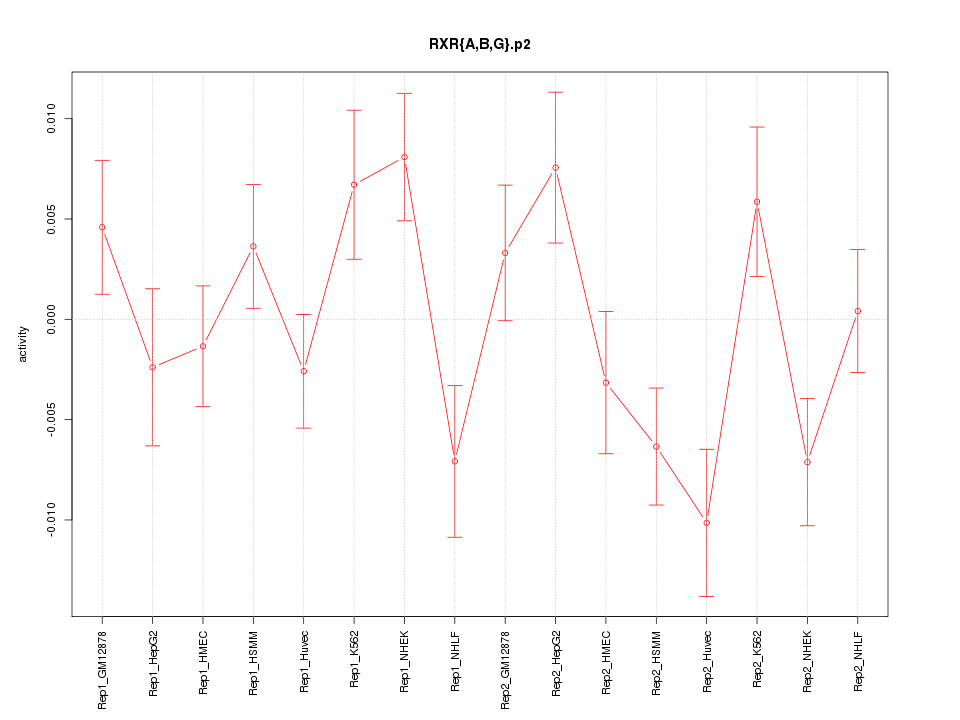
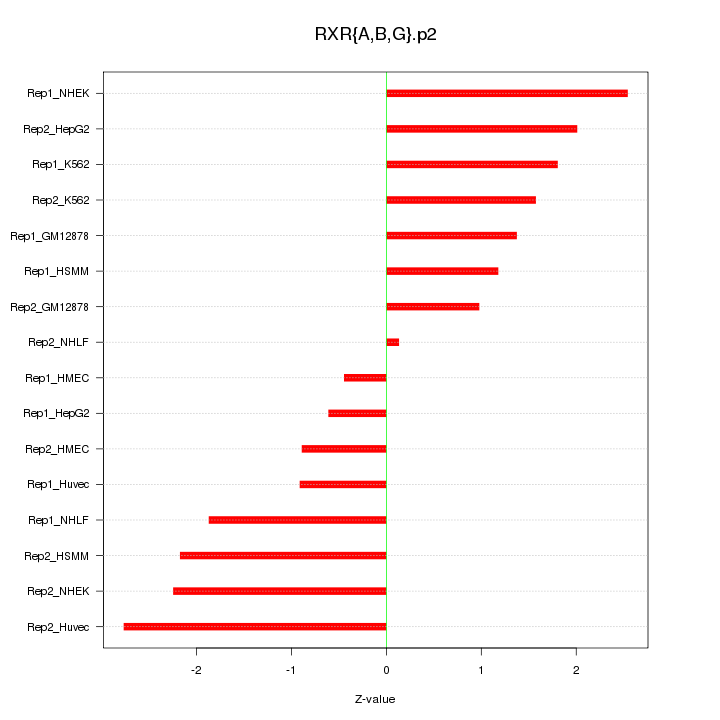
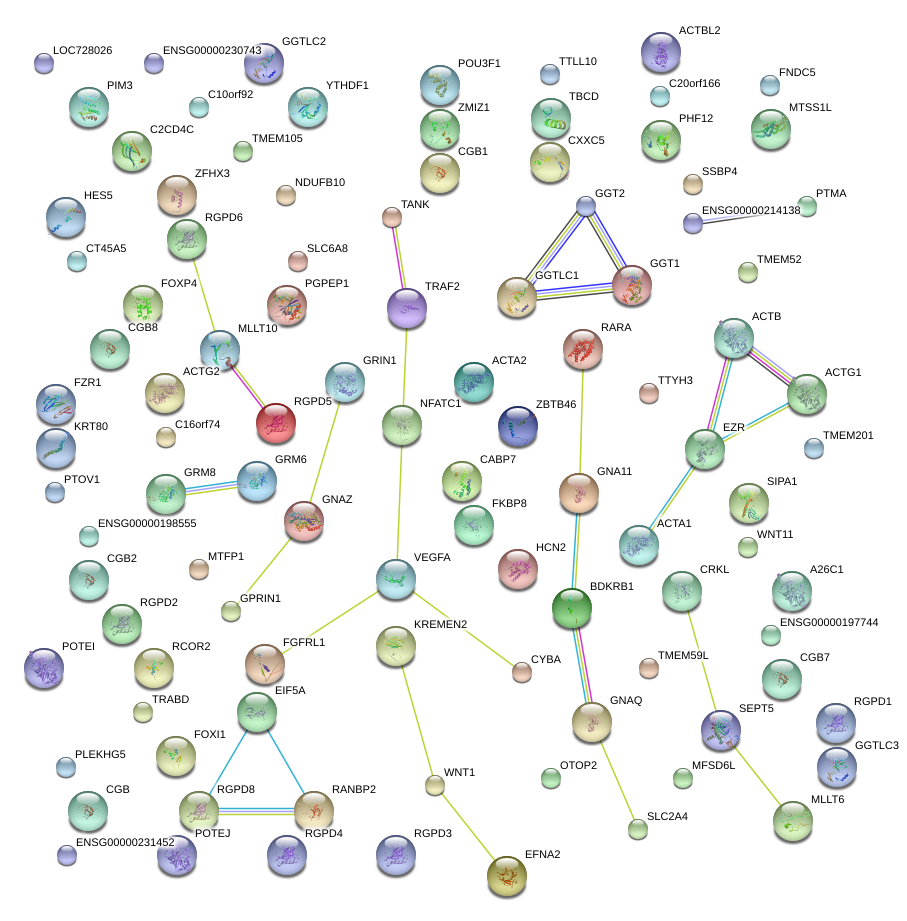
|
chr22_+_18081968
|
3.131
|
NM_002688
|
SEPT5
|
septin 5
|
|
chr6_+_43846372
|
2.604
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr6_+_43845915
|
2.188
|
NM_001025366
NM_001025367
NM_001025368
NM_001025369
NM_001025370
NM_001033756
NM_001171622
NM_001171623
NM_001171624
NM_001171625
NM_001171626
NM_001171627
NM_001171628
NM_001171629
NM_001171630
NM_003376
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr22_+_18085891
|
2.173
|
|
SEPT5
|
septin 5
|
|
chr22_+_18085990
|
2.131
|
|
SEPT5-GP1BB
SEPT5
|
SEPT5-GP1BB read-through transcript
septin 5
|
|
chr6_+_43845875
|
1.911
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr6_+_43846155
|
1.816
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr9_-_101621964
|
1.781
|
|
|
|
|
chr18_+_75261231
|
1.674
|
NM_172387
NM_172389
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr20_+_60558369
|
1.623
|
|
C20orf166
|
chromosome 20 open reading frame 166
|
|
chr6_+_43846735
|
1.621
|
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr20_-_61933012
|
1.611
|
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr1_+_9571518
|
1.526
|
NM_001010866
NM_001130924
|
TMEM201
|
transmembrane protein 201
|
|
chr22_+_48966394
|
1.509
|
|
TRABD
|
TraB domain containing
|
|
chr17_+_7125702
|
1.473
|
NM_001042
|
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4
|
|
chr11_-_75595098
|
1.397
|
NM_004626
|
WNT11
|
wingless-type MMTV integration site family, member 11
|
|
chr17_+_34115383
|
1.312
|
NM_005937
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr5_+_139040479
|
1.293
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr22_+_48966480
|
1.273
|
NM_025204
|
TRABD
|
TraB domain containing
|
|
chr1_-_38285033
|
1.266
|
NM_002699
|
POU3F1
|
POU class 3 homeobox 1
|
|
chr12_-_50871953
|
1.220
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chr16_-_84342129
|
1.219
|
NM_206967
|
C16orf74
|
chromosome 16 open reading frame 74
|
|
chr22_+_21742727
|
1.210
|
|
GNAZ
|
guanine nucleotide binding protein (G protein), alpha z polypeptide
|
|
chr17_+_35751796
|
1.195
|
NM_001024809
|
RARA
|
retinoic acid receptor, alpha
|
|
chr22_+_21742364
|
1.192
|
NM_002073
|
GNAZ
|
guanine nucleotide binding protein (G protein), alpha z polypeptide
|
|
chr22_+_28446288
|
1.186
|
NM_182527
|
CABP7
|
calcium binding protein 7
|
|
chr22_+_22703107
|
1.178
|
NM_001144931
|
LOC391322
|
D-dopachrome tautomerase-like
|
|
chrX_+_152606945
|
1.135
|
NM_001142805
NM_005629
|
SLC6A8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8
|
|
chr4_+_995609
|
1.127
|
NM_001004356
NM_001004358
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr19_+_1236766
|
1.124
|
NM_001405
|
EFNA2
|
ephrin-A2
|
|
chr2_+_232281491
|
1.086
|
|
PTMA
|
prothymosin, alpha
|
|
chr17_-_24302597
|
1.060
|
NM_001033561
NM_020889
|
PHF12
|
PHD finger protein 12
|
|
chr10_-_134605978
|
1.053
|
NM_173572
|
C10orf93
|
chromosome 10 open reading frame 93
|
|
chr4_+_995395
|
1.052
|
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr17_+_7151653
|
0.993
|
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr19_+_55046229
|
0.991
|
|
PTOV1
|
prostate tumor overexpressed 1
|
|
chr16_+_2954217
|
0.956
|
NM_024507
NM_172229
|
KREMEN2
|
kringle containing transmembrane protein 2
|
|
chr7_+_2638108
|
0.947
|
NM_025250
|
TTYH3
|
tweety homolog 3 (Drosophila)
|
|
chr11_+_65162114
|
0.945
|
NM_006747
|
SIPA1
|
signal-induced proliferation-associated 1
|
|
chr1_-_33108876
|
0.944
|
NM_001171940
NM_153756
|
FNDC5
|
fibronectin type III domain containing 5
|
|
chr8_-_142586487
|
0.928
|
NM_207414
|
FLJ43860
|
FLJ43860 protein
|
|
chr6_+_41622267
|
0.923
|
|
FOXP4
|
forkhead box P4
|
|
chr1_+_1104939
|
0.921
|
NM_153254
|
TTLL10
|
tubulin tyrosine ligase-like family, member 10
|
|
chr19_+_3045528
|
0.912
|
|
GNA11
|
guanine nucleotide binding protein (G protein), alpha 11 (Gq class)
|
|
chr2_+_113709553
|
0.911
|
|
LOC654433
|
hypothetical LOC654433
|
|
chr17_-_76985538
|
0.906
|
|
|
|
|
chr17_+_70431964
|
0.889
|
NM_178160
|
OTOP2
|
otopetrin 2
|
|
chr7_-_126679540
|
0.888
|
NM_001127323
|
GRM8
|
glutamate receptor, metabotropic 8
|
|
chr22_+_17539198
|
0.883
|
|
|
|
|
chr20_-_56859352
|
0.879
|
|
GNAS-AS1
|
GNAS antisense RNA 1 (non-protein coding)
|
|
chr1_-_1840564
|
0.876
|
NM_178545
|
TMEM52
|
transmembrane protein 52
|
|
chr17_-_24302450
|
0.866
|
|
PHF12
|
PHD finger protein 12
|
|
chr7_-_5534252
|
0.861
|
|
ACTB
|
actin, beta
|
|
chr22_+_48740432
|
0.841
|
|
PIM3
|
pim-3 oncogene
|
|
chr5_+_169465478
|
0.840
|
NM_012188
NM_144769
|
FOXI1
|
forkhead box I1
|
|
chr20_-_61317891
|
0.827
|
NM_017798
|
YTHDF1
|
YTH domain family, member 1
|
|
chr16_-_69277355
|
0.826
|
NM_138383
|
MTSS1L
|
metastasis suppressor 1-like
|
|
chr7_+_2638156
|
0.826
|
|
TTYH3
|
tweety homolog 3 (Drosophila)
|
|
chr19_+_18391211
|
0.825
|
NM_001009998
NM_032627
|
SSBP4
|
single stranded DNA binding protein 4
|
|
chr19_+_18391423
|
0.809
|
|
|
|
|
chr7_-_5534737
|
0.801
|
|
ACTB
|
actin, beta
|
|
chr19_+_3457264
|
0.793
|
NM_016263
|
FZR1
|
fizzy/cell division cycle 20 related 1 (Drosophila)
|
|
chr11_-_63440891
|
0.790
|
NM_173587
|
RCOR2
|
REST corepressor 2
|
|
chr9_+_138900760
|
0.788
|
NM_021138
|
TRAF2
|
TNF receptor-associated factor 2
|
|
chr19_+_55046088
|
0.787
|
NM_017432
|
PTOV1
|
prostate tumor overexpressed 1
|
|
chr10_+_80740584
|
0.784
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr12_+_47658502
|
0.784
|
NM_005430
|
WNT1
|
wingless-type MMTV integration site family, member 1
|
|
chr19_-_360138
|
0.766
|
NM_001136263
|
C2CD4C
|
C2 calcium-dependent domain containing 4C
|
|
chr1_-_2451543
|
0.761
|
NM_001010926
|
HES5
|
hairy and enhancer of split 5 (Drosophila)
|
|
chr6_+_33044353
|
0.748
|
|
|
|
|
chr22_+_29151651
|
0.746
|
NM_001003704
NM_016498
|
MTFP1
|
mitochondrial fission process 1
|
|
chr1_-_6468108
|
0.744
|
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr19_+_18312365
|
0.735
|
NM_017712
|
FKBP8
PGPEP1
|
FK506 binding protein 8, 38kDa
pyroglutamyl-peptidase I
|
|
chr16_-_71650034
|
0.729
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr17_-_8643163
|
0.727
|
NM_152599
|
MFSD6L
|
major facilitator superfamily domain containing 6-like
|
|
chr22_+_19601723
|
0.726
|
|
CRKL
|
v-crk sarcoma virus CT10 oncogene homolog (avian)-like
|
|
chr19_+_540849
|
0.724
|
NM_001194
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2
|
|
chr22_+_21318779
|
0.723
|
NM_199127
|
GGTLC2
|
gamma-glutamyltransferase light chain 2
|
|
chr19_-_55046117
|
0.719
|
|
LOC100506033
|
hypothetical LOC100506033
|
|
chr16_+_1949517
|
0.719
|
NM_004548
|
NDUFB10
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10, 22kDa
|
|
chr2_-_112907577
|
0.715
|
NM_001164463
|
RGPD8
RGPD5
|
RANBP2-like and GRIP domain containing 8
RANBP2-like and GRIP domain containing 5
|
|
chr19_+_18584666
|
0.711
|
NM_012109
|
TMEM59L
|
transmembrane protein 59-like
|
|
chr16_-_87244896
|
0.709
|
NM_000101
|
CYBA
|
cytochrome b-245, alpha polypeptide
|
|
chr9_+_139153409
|
0.709
|
NM_000832
NM_001185090
NM_001185091
NM_007327
NM_021569
|
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1
|
|
chr17_-_76919068
|
0.708
|
NM_178520
|
TMEM105
|
transmembrane protein 105
|
|
chr22_+_19601721
|
0.708
|
|
CRKL
|
v-crk sarcoma virus CT10 oncogene homolog (avian)-like
|
|
chr19_-_54250808
|
0.705
|
NM_033142
|
CGB7
|
chorionic gonadotropin, beta polypeptide 7
|
|
chr7_-_4965270
|
0.705
|
NM_001100600
NM_198403
|
MMD2
|
monocyte to macrophage differentiation-associated 2
|
|
chr19_+_54872220
|
0.698
|
|
PRMT1
|
protein arginine methyltransferase 1
|
|
chr14_-_104706159
|
0.695
|
NM_002226
NM_145159
|
JAG2
|
jagged 2
|
|
chr3_-_50304706
|
0.689
|
NM_006764
|
IFRD2
|
interferon-related developmental regulator 2
|
|
chr17_+_7151368
|
0.686
|
NM_001970
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr17_-_40401138
|
0.684
|
NM_006688
|
C1QL1
|
complement component 1, q subcomponent-like 1
|
|
chr12_+_110328126
|
0.683
|
NM_005475
|
SH2B3
|
SH2B adaptor protein 3
|
|
chr1_+_3597092
|
0.680
|
NM_001126240
NM_001126241
NM_001126242
|
TP73
|
tumor protein p73
|
|
chr1_+_27192719
|
0.679
|
NM_001013642
|
TRNP1
|
TMF1-regulated nuclear protein 1
|
|
chr9_+_132874324
|
0.679
|
NM_006059
|
LAMC3
|
laminin, gamma 3
|
|
chr22_-_17892001
|
0.678
|
|
CLDN5
|
claudin 5
|
|
chr16_+_87765661
|
0.676
|
NM_004933
|
CDH15
|
cadherin 15, type 1, M-cadherin (myotubule)
|
|
chr6_+_30018287
|
0.673
|
NM_002116
|
HLA-A
|
major histocompatibility complex, class I, A
|
|
chr19_+_54872304
|
0.673
|
NM_001536
NM_198318
|
PRMT1
|
protein arginine methyltransferase 1
|
|
chr6_+_41712907
|
0.672
|
|
MDFI
|
MyoD family inhibitor
|
|
chr16_+_2138608
|
0.666
|
NM_014353
|
RAB26
|
RAB26, member RAS oncogene family
|
|
chr19_+_7874701
|
0.666
|
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr17_+_70343659
|
0.655
|
|
TMEM104
|
transmembrane protein 104
|
|
chr11_+_440260
|
0.655
|
NM_030783
|
PTDSS2
|
phosphatidylserine synthase 2
|
|
chr11_+_440346
|
0.653
|
|
PTDSS2
|
phosphatidylserine synthase 2
|
|
chr10_+_73703682
|
0.653
|
NM_019058
|
DDIT4
|
DNA-damage-inducible transcript 4
|
|
chr16_+_658158
|
0.653
|
|
RHOT2
|
ras homolog gene family, member T2
|
|
chr7_+_98994363
|
0.652
|
NM_001009958
NM_001009960
NM_001085367
NM_001085368
|
ZNF655
|
zinc finger protein 655
|
|
chr20_-_55718351
|
0.652
|
NM_020182
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr21_+_45649719
|
0.652
|
|
|
|
|
chr14_-_104705753
|
0.650
|
|
JAG2
|
jagged 2
|
|
chr22_-_17891777
|
0.650
|
|
CLDN5
|
claudin 5
|
|
chr17_+_45993338
|
0.649
|
NM_018896
NM_198376
NM_198377
NM_198378
NM_198379
NM_198380
NM_198382
NM_198383
NM_198384
NM_198385
NM_198386
NM_198387
NM_198388
NM_198396
NM_198397
|
CACNA1G
|
calcium channel, voltage-dependent, T type, alpha 1G subunit
|
|
chr7_+_100156358
|
0.648
|
NM_000799
|
EPO
|
erythropoietin
|
|
chr5_+_176786279
|
0.647
|
NM_001004105
NM_001004106
NM_002082
|
GRK6
|
G protein-coupled receptor kinase 6
|
|
chr11_-_45896144
|
0.643
|
NM_004813
NM_057174
|
PEX16
|
peroxisomal biogenesis factor 16
|
|
chr1_-_47468029
|
0.637
|
NM_003189
|
TAL1
|
T-cell acute lymphocytic leukemia 1
|
|
chr11_+_551449
|
0.636
|
NM_001143994
NM_003475
|
RASSF7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7
|
|
chr10_-_71002998
|
0.633
|
NM_020999
|
NEUROG3
|
neurogenin 3
|
|
chr4_-_2336328
|
0.633
|
NM_001172659
NM_001172660
|
ZFYVE28
|
zinc finger, FYVE domain containing 28
|
|
chr9_+_37640996
|
0.630
|
NM_014907
|
FRMPD1
|
FERM and PDZ domain containing 1
|
|
chr22_+_36384647
|
0.630
|
NM_020315
|
PDXP
|
pyridoxal (pyridoxine, vitamin B6) phosphatase
|
|
chr4_+_996251
|
0.629
|
NM_021923
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr17_-_45562112
|
0.628
|
NM_174920
|
SAMD14
|
sterile alpha motif domain containing 14
|
|
chr22_+_18485145
|
0.627
|
|
RANBP1
|
RAN binding protein 1
|
|
chr19_+_7874764
|
0.624
|
NM_145185
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr20_+_62158882
|
0.624
|
NM_198723
|
TCEA2
|
transcription elongation factor A (SII), 2
|
|
chr1_+_46541913
|
0.623
|
NM_006004
|
UQCRH
|
ubiquinol-cytochrome c reductase hinge protein
|
|
chr5_+_139040242
|
0.622
|
|
CXXC5
|
CXXC finger protein 5
|
|
chr6_+_31059463
|
0.621
|
NM_001010909
|
MUC21
|
mucin 21, cell surface associated
|
|
chr4_-_8180278
|
0.618
|
|
ABLIM2
|
actin binding LIM protein family, member 2
|
|
chr1_+_158317967
|
0.615
|
NM_004983
|
KCNJ9
|
potassium inwardly-rectifying channel, subfamily J, member 9
|
|
chr16_+_658081
|
0.612
|
NM_138769
|
RHOT2
|
ras homolog gene family, member T2
|
|
chr1_-_46541595
|
0.611
|
|
LRRC41
|
leucine rich repeat containing 41
|
|
chr19_+_44308224
|
0.609
|
NM_001014831
NM_001014832
NM_001014834
NM_001014835
NM_005884
|
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4
|
|
chr16_+_29725665
|
0.609
|
|
|
|
|
chr22_+_20068039
|
0.609
|
NM_001128635
|
RIMBP3B
|
RIMS binding protein 3B
|
|
chr16_+_68765428
|
0.606
|
NM_173619
|
CLEC18C
CLEC18A
CLEC18B
|
C-type lectin domain family 18, member C
C-type lectin domain family 18, member A
C-type lectin domain family 18, member B
|
|
chr4_-_154929657
|
0.606
|
NM_003013
|
SFRP2
|
secreted frizzled-related protein 2
|
|
chr1_-_46541560
|
0.606
|
|
|
|
|
chr19_+_935316
|
0.605
|
NM_024100
|
WDR18
|
WD repeat domain 18
|
|
chr22_+_18485095
|
0.604
|
|
RANBP1
|
RAN binding protein 1
|
|
chr6_+_31731647
|
0.603
|
NM_019101
|
APOM
|
apolipoprotein M
|
|
chr16_+_22733307
|
0.602
|
NM_006043
|
HS3ST2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2
|
|
chr16_+_1299674
|
0.602
|
|
UBE2I
|
ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast)
|
|
chr6_-_150431864
|
0.601
|
NM_024518
|
ULBP3
|
UL16 binding protein 3
|
|
chr19_+_17047569
|
0.600
|
NM_001130065
NM_004145
|
MYO9B
|
myosin IXB
|
|
chr19_+_7874765
|
0.598
|
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr1_-_46541525
|
0.595
|
|
LRRC41
|
leucine rich repeat containing 41
|
|
chr2_+_111206620
|
0.595
|
NM_001142807
|
ACOXL
|
acyl-CoA oxidase-like
|
|
chr10_-_80875097
|
0.593
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr7_-_150283794
|
0.593
|
NM_172057
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr11_+_67563093
|
0.591
|
|
TCIRG1
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3
|
|
chr1_-_46541618
|
0.590
|
NM_006369
|
LRRC41
|
leucine rich repeat containing 41
|
|
chr11_+_440408
|
0.588
|
|
PTDSS2
|
phosphatidylserine synthase 2
|
|
chr17_+_35046843
|
0.583
|
NM_001165937
NM_001165938
NM_006804
|
STARD3
|
StAR-related lipid transfer (START) domain containing 3
|
|
chr20_+_54637714
|
0.579
|
NM_003222
|
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma)
|
|
chr6_+_30018265
|
0.579
|
|
HLA-A
|
major histocompatibility complex, class I, A
|
|
chr3_+_51680261
|
0.578
|
NM_001129884
NM_015926
|
TEX264
|
testis expressed 264
|
|
chr1_+_200584421
|
0.576
|
NM_001167857
NM_001167858
NM_002481
|
PPP1R12B
|
protein phosphatase 1, regulatory (inhibitor) subunit 12B
|
|
chr6_-_3172775
|
0.575
|
NM_178012
|
TUBB2B
|
tubulin, beta 2B
|
|
chr12_-_55920741
|
0.574
|
NM_020142
|
NDUFA4L2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
|
|
chr19_+_7874762
|
0.573
|
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr19_-_55046718
|
0.569
|
|
LOC100506033
|
hypothetical LOC100506033
|
|
chr19_+_7874774
|
0.568
|
|
MAP2K7
|
mitogen-activated protein kinase kinase 7
|
|
chr5_-_746413
|
0.568
|
NM_007030
|
TPPP
|
tubulin polymerization promoting protein
|
|
chr19_+_52451570
|
0.561
|
NM_015603
|
CCDC9
|
coiled-coil domain containing 9
|
|
chr6_+_31731667
|
0.561
|
|
APOM
|
apolipoprotein M
|
|
chr19_-_9908014
|
0.560
|
NM_058164
|
OLFM2
|
olfactomedin 2
|
|
chr19_+_1389410
|
0.559
|
|
RPS15
|
ribosomal protein S15
|
|
chr17_+_8132539
|
0.558
|
NM_001177801
NM_001177802
NM_016492
|
RANGRF
|
RAN guanine nucleotide release factor
|
|
chr11_+_72765318
|
0.557
|
NM_032871
|
RELT
|
RELT tumor necrosis factor receptor
|
|
chr16_+_1299693
|
0.555
|
|
UBE2I
|
ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast)
|
|
chr1_-_201411533
|
0.550
|
NM_004997
|
MYBPH
|
myosin binding protein H
|
|
chr12_+_88268845
|
0.547
|
|
LOC100131490
|
hypothetical LOC100131490
|
|
chr2_+_220200530
|
0.547
|
NM_005070
NM_201574
|
SLC4A3
|
solute carrier family 4, anion exchanger, member 3
|
|
chr11_+_817584
|
0.544
|
NM_173584
|
EFCAB4A
|
EF-hand calcium binding domain 4A
|
|
chr16_-_29818334
|
0.544
|
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2
|
|
chr20_+_44070946
|
0.542
|
NM_004994
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase)
|
|
chr4_-_658035
|
0.542
|
NM_007100
|
ATP5I
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit E
|
|
chr19_-_18409971
|
0.541
|
NM_001170938
NM_016368
|
ISYNA1
|
inositol-3-phosphate synthase 1
|
|
chr19_-_16494897
|
0.535
|
|
CHERP
|
calcium homeostasis endoplasmic reticulum protein
|
|
chr20_+_43996717
|
0.535
|
NM_022104
|
PCIF1
|
PDX1 C-terminal inhibiting factor 1
|
|
chr17_+_71097147
|
0.530
|
|
MYO15B
|
myosin XVB pseudogene
|
|
chr11_-_62136778
|
0.530
|
NM_153265
|
EML3
|
echinoderm microtubule associated protein like 3
|
|
chr16_+_2145810
|
0.528
|
|
TRAF7
|
TNF receptor-associated factor 7
|
|
chr9_-_130006309
|
0.528
|
NM_001131015
NM_012127
|
CIZ1
|
CDKN1A interacting zinc finger protein 1
|
|
chr1_+_205137410
|
0.527
|
NM_001185156
NM_001185157
NM_001185158
NM_006850
|
IL24
|
interleukin 24
|
|
chr19_+_17691302
|
0.524
|
NM_018174
|
MAP1S
|
microtubule-associated protein 1S
|
|
chr20_+_62164900
|
0.524
|
|
TCEA2
|
transcription elongation factor A (SII), 2
|
|
chr11_+_43920385
|
0.523
|
NM_001145033
|
C11orf96
|
chromosome 11 open reading frame 96
|
|
chr19_+_55579272
|
0.523
|
|
POLD1
|
polymerase (DNA directed), delta 1, catalytic subunit 125kDa
|
|
chr16_+_1299654
|
0.522
|
|
UBE2I
|
ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast)
|
|
chr16_+_1299637
|
0.520
|
NM_003345
NM_194259
|
UBE2I
|
ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast)
|
|
chr13_+_113369470
|
0.520
|
NM_002929
|
GRK1
|
G protein-coupled receptor kinase 1
|
|
chr2_+_95326753
|
0.520
|
NM_013434
|
KCNIP3
|
Kv channel interacting protein 3, calsenilin
|
|
chr19_+_52451642
|
0.519
|
|
CCDC9
|
coiled-coil domain containing 9
|
|
chr5_+_139040370
|
0.518
|
|
CXXC5
|
CXXC finger protein 5
|